|
|
Home
Pearsall Family DNA
Surname Project
Number of Pearsalls
By Location
Maps by Family
Surname
The History of the
Parshall Family from the Conquest of England by William of Normandy, A.D.
1066 to the Close of the 19th Century (1903)
The Parshall Family
A.D. 870-1913 (1915)
History and
Genealogy of the Pearsall Family in England and America (1928)
Volume I
Volume II
Volume III
|
|
The Pearsall Family DNA – Surname
Project
RESULTS
As this family DNA surname project
develops, more information for family member results will be posted.
Individuals will not be identified by name to protect privacy; we will
use surname, location, and a code.
123-MARKER RESULTS
Different labs test different Y-DNA
markers. DNA Heritage and Relative Genetics use the same 43-marker
standards (SGMF), these two labs are highly recommended for this project.
FamilyTreeDNA, Ethnoancestry,
DNA-Fingerprint, and GeneBase have a variety of other markers optionally
available for academic purposes.
FamilyTreeDNA/DNA-Fingerprint provides DNA storage (which maybe a
consideration when testing elderly family members), as well as
autosomal/CODIS markers, and mtDNA testing.
The 123-marker test results for myself
are below:
|
Name
|
Pearsall,
James A.
|
|
Haplogroup,
Haplotype
|
I1b1* –
Confirmed
|
|
DXYS156-Y
|
12
|
|
DYF371a
|
10c
|
|
DYF371b
|
12t
|
|
DYF371c
|
13c
|
|
DYF371d
|
14c
|
|
DYF385 S1 a
|
10
|
|
DYF385 S1 b
|
10
|
|
DYF397a
|
13
|
|
DYF397b
|
13
|
|
DYF397c
|
14
|
|
DYF397d
|
14
|
|
DYF399 S1 a
|
20c
|
|
DYF399 S1 b
|
21t
|
|
DYF399 S1 c
|
23c
|
|
DYF401 S1 a (DYS527)
|
18
|
|
DYF401 S1 b
|
18
|
|
DYF406 S1
|
5
|
|
DYF408a
|
188
|
|
DYF408b
|
188
|
|
DYF408c
|
9
|
|
DYF408d
|
9
|
|
DYF411a
|
12
|
|
DYF411b
|
14
|
|
DYS19 (DYS394)
|
15
|
|
DYS385a
|
12
|
|
DYS385b
|
15
|
|
DYS388
|
15
|
|
DYS389I
|
14
|
|
DYS389II
|
30
|
|
DYS390
|
23
|
|
DYS391
|
10
|
|
DYS392
|
11
|
|
DYS393
|
13
|
|
DYS395S1a
|
16
|
|
DYS395S1b
|
17
|
|
DYS406S1
|
12
|
|
DYS413a
|
21
|
|
DYS413b
|
21
|
|
DYS425 (DYF371t)
|
12
|
|
DYS426
|
11
|
|
DYS434
|
10
|
|
DYS435
|
11
|
|
DYS436
|
12
|
|
DYS437
|
14
|
|
DYS438
|
10
|
|
DYS439 (Y-GATA-A4)
|
11
|
|
DYS441
|
15 *SMGF/RG/DNAH std
14 *GB std
|
|
DYS442
|
12
|
|
DYS444
|
14
|
|
DYS445
|
11
|
|
DYS446
|
11 *SMGF/RG/DNAH std
11 *FTDNA std
12 *GB std
|
|
DYS447
|
25
|
|
DYS448
|
18
|
|
DYS449
|
28
|
|
DYS450
|
8
|
|
DYS452
|
11
|
|
DYS453
|
10
|
|
DYS454
|
11
|
|
DYS455
|
11 *SMGF/RG/DNAH std
11 *FTDNA std
10 *GB std
|
|
DYS456
|
14
|
|
DYS458
|
17 *SMGF/RG/DNAH std
17 *FTDNA std
15 *GB std
|
|
DYS459a
|
8
|
|
DYS459b
|
9
|
|
DYS460 (Y-GATA-A7-1)
|
10
|
|
DYS461 (Y-GATA-A7-2)
|
12
|
|
DYS462
|
12
|
|
DYS463
|
20
|
|
DYS464a
|
11g
|
|
DYS464b
|
14g
|
|
DYS464c
|
14g
|
|
DYS464d
|
15g
|
|
DYS472
|
8
|
|
DYS481
|
23 *EA std
27 *FTDNA std
|
|
DYS485
|
15
|
|
DYS487
|
12
|
|
DYS490
|
11 *EA std
12 *FTDNA std
|
|
DYS492
|
12
|
|
DYS494
|
10
|
|
DYS495
|
15
|
|
DYS505
|
11
|
|
DYS511
|
10
|
|
DYS520
|
20
|
|
DYS522
|
11 *EA std
10 *GB std
|
|
DYS531
|
12 *EA std
11 *FTDNA std
10 *GB std
|
|
DYS533
|
12
|
|
DYS534
|
15
|
|
DYS537
|
11
|
|
DYS549
|
12
|
|
DYS556
|
11
|
|
DYS557
|
18
|
|
DYS565
|
11
|
|
DYS568
|
12
|
|
DYS570
|
18
|
|
DYS572
|
10
|
|
DYS575
|
10
|
|
DYS576
|
17
|
|
DYS578
|
8
|
|
DYS589
|
1
|
|
DYS590
|
8
|
|
DYS594
|
10
|
|
DYS607
|
10
|
|
DYS617
|
14
|
|
DYS635 (Y-GATA-C4)
|
21
|
|
DYS636
|
10
|
|
DYS638
|
11
|
|
DYS640
|
11
|
|
DYS641
|
10
|
|
DYS643
|
10
|
|
DYS714
|
26
|
|
DYS716
|
27
|
|
DYS717
|
19
|
|
DYS724a (CDY a)
|
34
|
|
DYS724b (CDY b)
|
35
|
|
DYS725a
|
30
|
|
DYS725b
|
31
|
|
DYS725c
|
31
|
|
DYS725d
|
31
|
|
DYS726
|
12
|
|
YCA II a
|
21
|
|
YCA II b
|
21
|
|
Y-GATA-A10
|
14 *SMGF/RG/DNAH std
16 *GB std
|
|
Y-GATA-H4 (TAGA)
|
11 *SMGF/RG/DNAH std
11 *GB std
10 *FTDNA std
|
|
Y-GGAAT1-B07
|
11
|
|
YAP
|
n/a
|
|
|
|
|
Deep SNP-I
Results
|
I1b1* -
Confirmed
|
|
M89 (F-R) – DNAH
|
Positive (+)
|
|
M168 (C-R) – DNAH
|
Positive (+)
|
|
M170 (I) – EA
|
Derived (+)
|
|
M258 (I) – FTDNA
|
Positive (+)
|
|
P19 (I) – FTDNA
|
Positive (+)
|
|
P37.2 (I1b) – EA/FTDNA
|
Derived (+)
|
|
P38 (I1) – FTDNA
|
Positive (+)
|
|
S31 (I*) – EA
|
Derived (+)
|
|
|
|
|
M9 (K-R) – DNAH
|
Negative (-)
|
|
M21 (I1a2) – FTDNA
|
Negative (-)
|
|
M26 (I1b2) –
DNAH
|
Negative (-)
|
|
M45 (P-R) – DNAH
|
Negative (-)
|
|
M52 (H) – DNAH
|
Negative (-)
|
|
M72 (I1a3) – FTDNA
|
Negative (-)
|
|
M96 (E) – DNAH
|
Negative (-)
|
|
M122 (O3) – DNAH
|
Negative (-)
|
|
M161 (I1b2a) – FTDNA
|
Negative (-)
|
|
M175 (O) – DNAH
|
Negative (-); expected deletion – likely reporting error
|
|
M201 (G) – DNAH
|
Negative (-)
|
|
M207 (R) – DNAH
|
Negative (-)
|
|
M223 (I1c) – FTDNA
|
Negative (-)
|
|
M227 (I1a4) – FTDNA
|
Negative (-)
|
|
M253 (I1a) –
DNAH
|
Negative (-)
|
|
M304 (J) – DNAH
|
Negative (-)
|
|
M307 (I1a) – FTDNA
|
Negative (-)
|
|
M343 (R1c) – DNAH
|
Negative (-)
|
|
P15 (G2) – DNAH
|
Negative (-)
|
|
P16 (G2a) – DNAH
|
Negative (-)
|
|
P30 (I1a) – FTDNA
|
Negative (-)
|
|
P37.2 (I1b) – DNAH
|
Negative (-); likely
lab error
|
|
P41.2 (I1b1b) – EA
|
Ancestral (-)
|
|
S23 (I1b2) – EA
|
Ancestral (-)
|
|
|
|
|
|
|
|
|
|
|
highlight –
conflicting results (working to resolve)
|
|
bold – FTDNA
also to test
|
|
GB – conversion to others not currently known
|
|
|
|

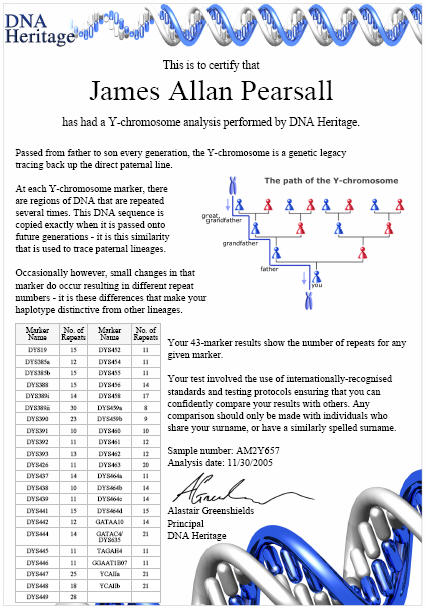

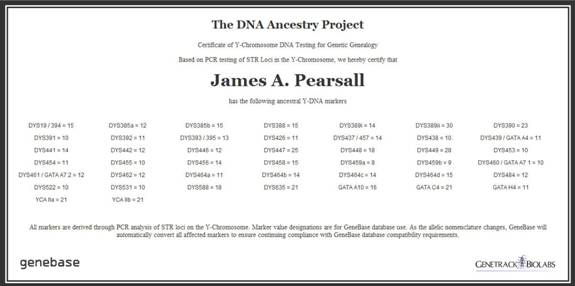


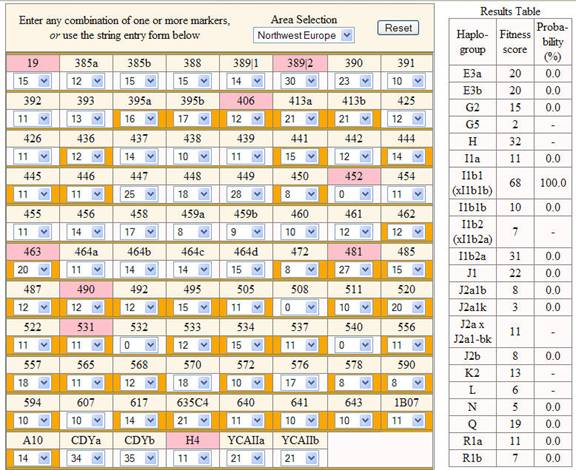
Y-DNA (STR) results also listed with the following public
databases:
Ybase.org
(CWWL6)
Ymatch.org
(pearsallj)
Ysearch.org
(28F42)
TO INITIAL PROJECT RESULTS (updated
August 23, 2006)--à
|
|